Some time ago I wrote a review of MOE the molecular modelling package from Chemical Computing Group, recently CCG announced and update and I thought it would be worth having a look at the new features. There are also a couple of things that are minor updates that will make my life easier, the scroll wheel on my mouse is now supported and there are alternating row background colours in the table view which helps when you have large numbers of columns.
The release notes highlight a number of new features.
- SAReport
- New or updated tools for manipulating sd files
- sdfrag
- sdstereo
- sdflter
- sdwash
- sdfilter
- BREED
- Improvements to surfaces and maps
- Protein Sidechain Conformation Library
- Small Molecule Scaffold Database
- Ray-tracer output.
- MOEvie Maker
Of these SAReport seems the most interesting
SAReport
SAReport is a new web application that analyzes 2D collections of project compounds and produces a Browser-based Structure-Activity Report in HTML format. The input data is typically a collection of lead candidate structures, ideally consisting of between 100 and 1000 molecules, I tested this using 100 structures with a single pIC50 for each compound and found it took several minutes on my MacBook Pro. Detection of common scaffolds can be done automatically, or the user can supply suggestions as shown below.
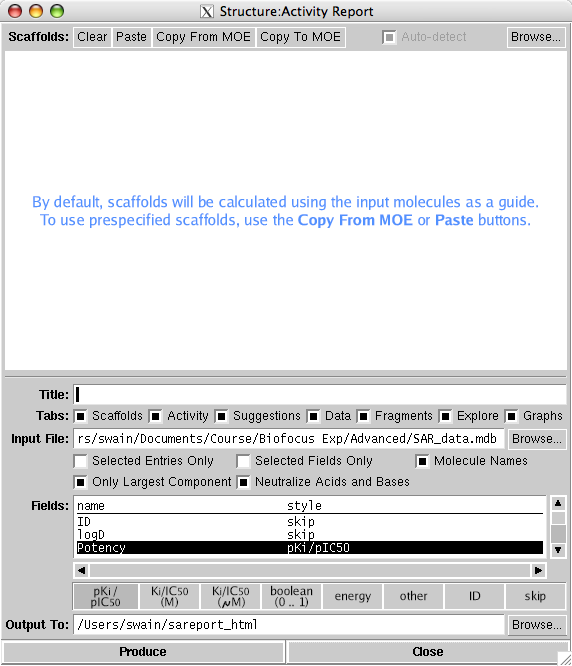
Correlation and assignment of molecules to multiple possible scaffolds is achieved by an algorithm which can operate with or without guidance from the operator. Specialized depiction methods are used to present scaffolds and R-groups. The HTML page which is generated by the report application contains a number of tabs for examining the common scaffolds, R-groups, structure vs. activity, fragmentation, and various graphs and interactive widgets. Perhaps the most important feature is the output can be viewed independently of MOE and so can be viewed and shared with other chemists. However any major changes to the content of the report such as adding molecules or modifying the scaffolds requires rebuilding the page with MOE.
The output is shown below
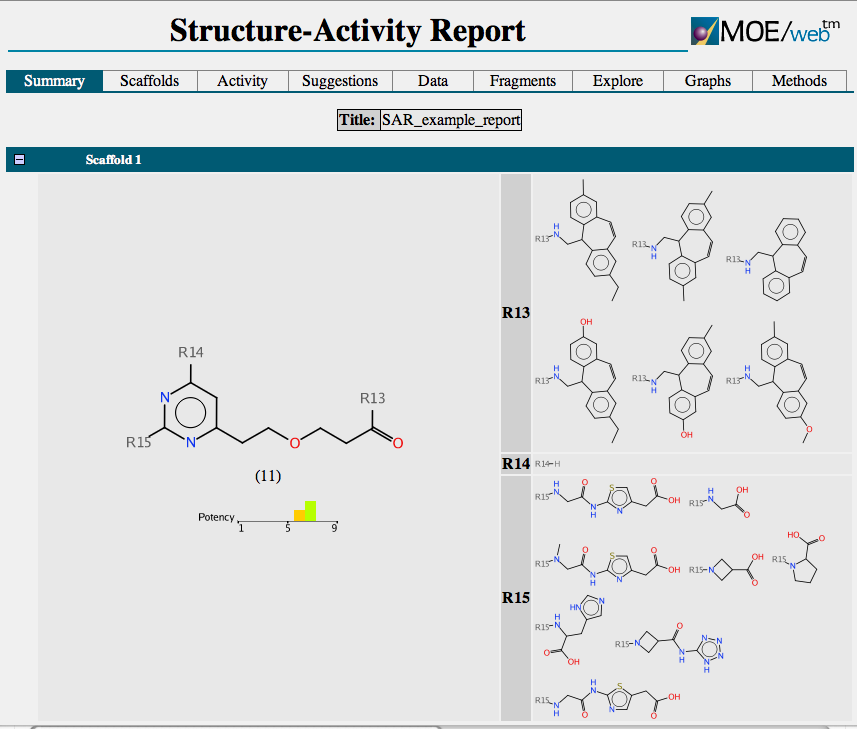
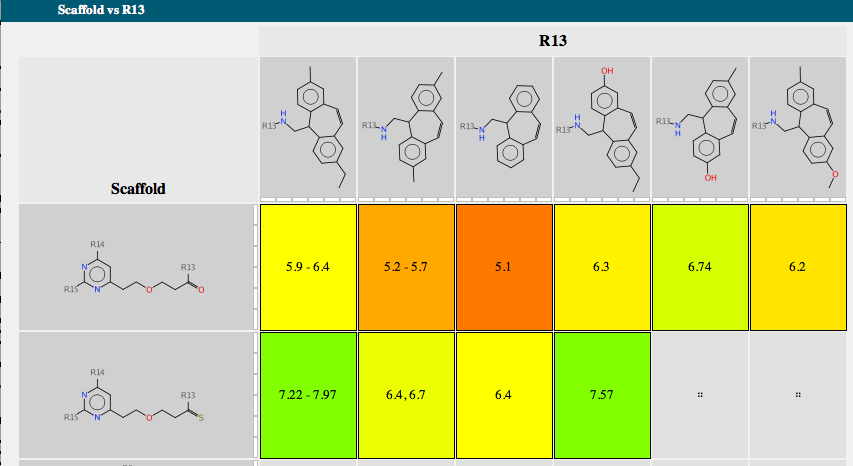
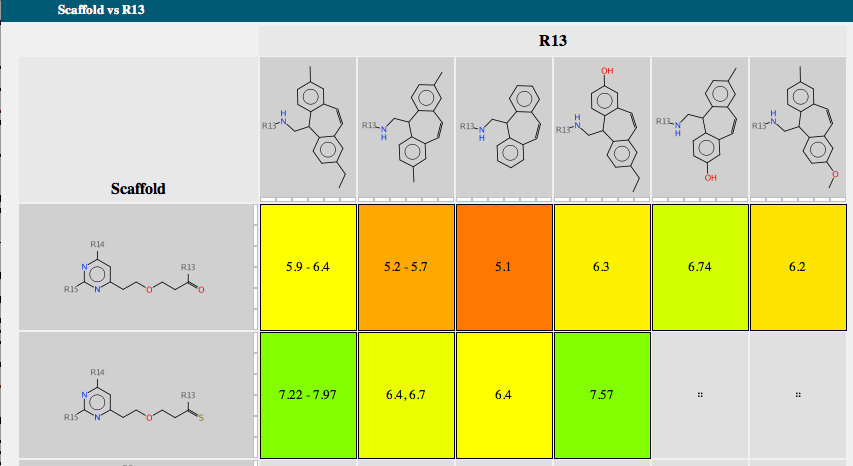
Each scaffold is shown together with a bar chart showing the range of activity and a list of R-groups. If you mouse over one of the R-groups a popup window displays all the molecules containing that R-group with the biological activity. A series of tabs across the top of the document gives access to a number of features. “Activity” allows the user to compare scaffold versus R-group or R-group versus R-group. The scaffolds are automatically aligned to ensure R-groups are mapped correctly.
The “Suggestions” tab is interesting, the structures shown in this tab are hypothetical molecules, constructed from motifs found in the input data, with statistical estimates of probable activity. I found it difficult to judge how effective this might be, but if nothing else it might highlight holes in the SAR.
The “Explore” tab allows the use limited ability to explore the data set. The selected molecule is positioned at the center of the cross-hairs. Each other molecule with data for the selected activity is plotted, above or below the current molecule depending on whether the activity is greater or less. Molecules plotted to the left have the same scaffold, and the distance indicates the extent to which the R-groups are different. Molecules plotted to the right have a different scaffold, or no scaffold, and the distance is proportional the the scaffold dissimilarity. Similarity is computed using MACCS fingerprints. If you hover over a point the structure and its activity is displayed, if you click on the point the graph is redrawn with the selected moleculae at the center.
This looks to be an interesting application for sharing information, and for the first iteration seems well designed, it will be interesting to see how much extra functionality will be added in the future.
SD tools
The SD Tools are a collection of commandline tools that are very useful for manipulating sd files. They are particularly useful for cleaning up sd files from commecial suppliers that don’t always have the correct/consistent ionisation states or have abbreviations for functional or protecting groups. sdfrag is designed to aid the fragmentation of molecular structures. I found it really useful when I was designing a fragment library recently.
BREED
BREED is an application for hybridizing a collection of aligned molecules has been added. The new application is based on Pierce’s program of the same name (BREED: Generating novel inhibitors through hybridization of known ligands. Application to CDK2, p38, and HIV protease.). New molecules are created from the aligned molecules by exchanging substituents along bonds that overlay in 3D. All such hybridizations are performed and the unique novel molecules are written to an output database.
The new ability to export as POV-Ray or movies will be an important addition for anyone who has to give presentations on machines that do not have access to MOE.
All in all an excellent update, some really interesting new applications and useful improvements on some of the existing tools.