infiniSee from BioSoveIT enables similarity searching, based on a query molecule, through billions of compounds in chemical spaces. Currently two chemical spaces from commercial compound vendors can be explored: the well-known Enamine REAL Space with 13 billion compounds and WuXi’s GalaXi with 1,7 billion compounds. The big advantage of these spaces is that any search hits can simply be ordered. The GUI is extremely easy to use – perfectly in line with the other well-known software platform (SeeSAR) by BioSolveIT. Behind the scenes infiniSee employs the FTrees similarity engine. FTrees captures a molecule’s topology and pharmacophore properties as a so-called Feature Tree. This enables finding compounds, which are structurally quite different (i.e. low Tanimoto similarity) but they have high pharmacophoric similarity, i.e. scaffold-hops that would normally not have been found.
A feature tree represents pharmacophoric fragments and functional groups of the molecule and the way these groups are linked together. Each node in the tree is labeled with a set of features representing chemical properties of the part of the molecule corresponding to the node. The comparison of feature trees is based on matching subtrees of two feature trees onto each other.
One major advantage of this technique is that a combinatorial approach is applied, molecules are built on the fly and chemical spaces can be explored directly, without explicit enumeration. (J Comput Aided Mol Des. 2001 Jun; 15(6): 497–520) DOI. This makes searching billions of compounds possible within minutes. A more recent enhancement is that the user can also determine more important parts of the query, to which hits should be highly similar.
From download to hit molecules
The download and installation are straightforward and infiniSee runs on Linux, Windows as well as Apple’s Mac OS X (see https://www.biosolveit.de/download/?product=infinisee). After the installation, you download the spaces desired for searching (see https://www.biosolveit.de/CoLibri/spaces.html). You then load your query molecule as sdf-, smiles or mol2 file format. A click on the green play-button then executes the search, which usually takes only minutes to complete. Try infiniSee for free for the first few days after downloading and installing. If you then need more time, simply request a trial license.
All searches take place behind your firewall, so in complete secrecy. Results can easily be selected and exported to file. Such file can then simply be mailed to the respective compound vendor requesting a quote. Typical synthesis times are a few weeks and success rates are claimed to be 70 – 80% or higher. BioSolveIT has made it possible for you to make a leap in your drug design project. Following our mantra: “fast, visual and easy”! Read on for a more in-depth intro.
Step-by-step introduction
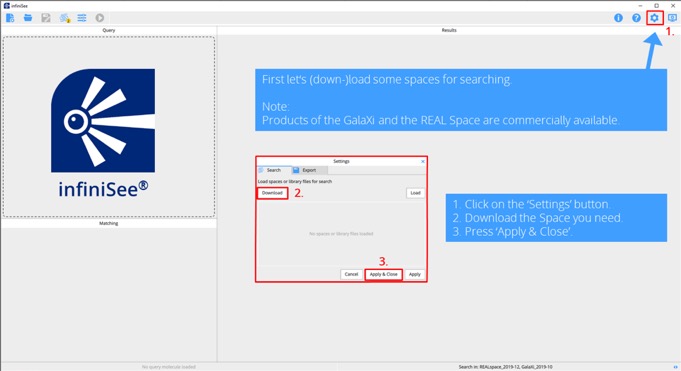
Step 1. Open infiniSee and download the chemical spaces or load your own space.
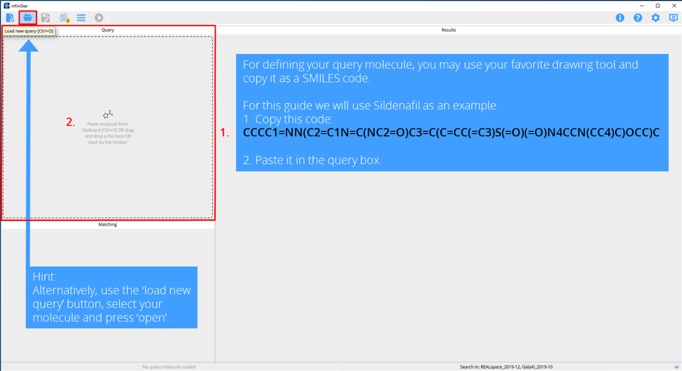
Step 2. Add your query molecule.
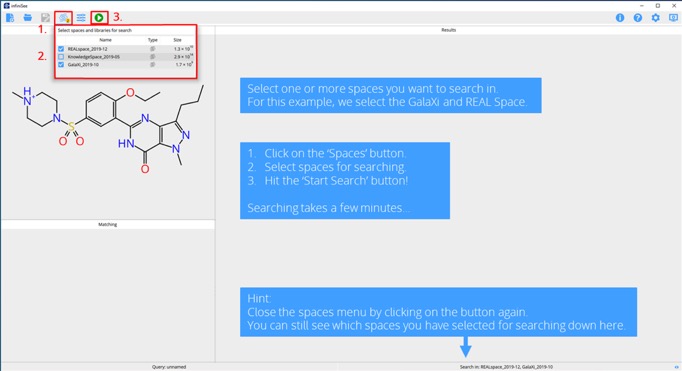
Step 3. Select the spaces and start searching.
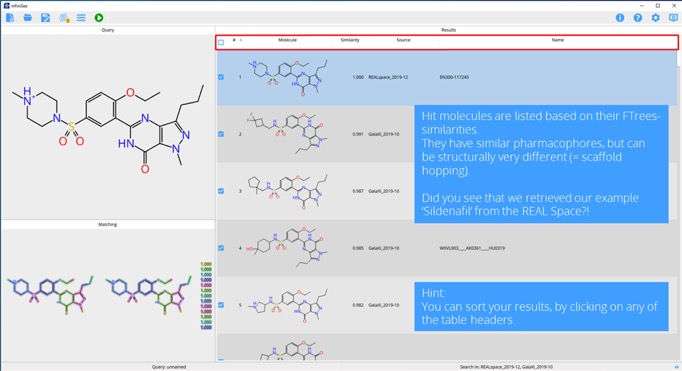
Step 4. Click on any entry in the results table and compare local similarities.
tep 5. Refine your searches.
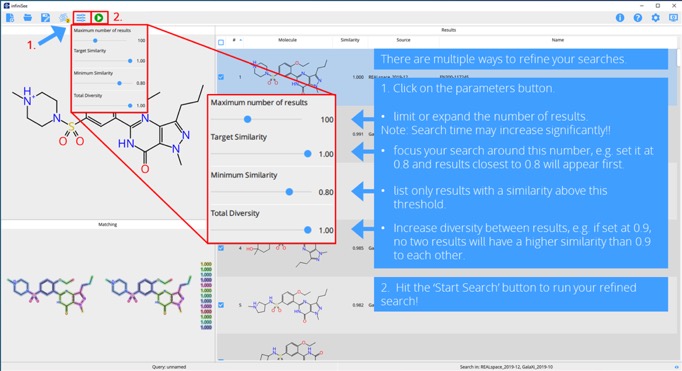
Step 6. Refine by determining important parts of your query.
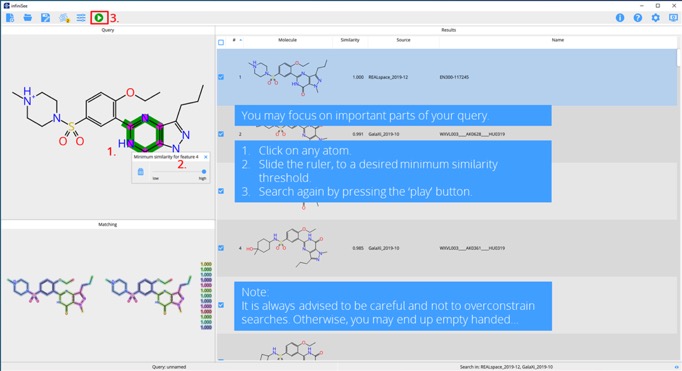
Results can easily be selected and then exported to file, it can be very useful to use a complementary virtual screening approach such as docking to further refine the results.
Last updated 13 January 2020