A recent publication described CypReact: A Software Tool for in Silico Reactant Prediction for Human Cytochrome P450 Enzymes DOI. There are a number of tools that predict sites of metabolism on a molecule and I’ve mentioned a couple FAME and SMARTCyp in the past.
Whilst the above programs offer a nice insight into the potential soft spots in the molecule sometimes you just want to know which enzymes are likely to be involved in the metabolism of a molecule, CypReact DOItakes a structure (SMILES or sdf input) and predicts if the molecule will react with any one of the nine of the most important human cytochrome P450 (CYP450) enzymes [CYP1A2, CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP2E1, or CYP3A4]. CypReact is a command line tool, is extremely fast and is ideal for quickly evaluating a batch of compounds. CypReact is available for download at https://bitbucket.org/Leon_Ti/cypreact.
The user can either input a .sdf file or a .csv. If the user input a .csv file, it should contain the SMILEs of all molecules and split them with “,”. The user can output a .sdf file or a .csv file. The format for the command is (using full path names).
|
1 2 |
<br>MacPro:~ Chris$ java -jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/cypreact.jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/ /Users/Chris/Desktop/SampleFiles/emend.sdf /Users/Chris/Desktop/SampleFiles/emendmetab.sdf 1A2,2A6,2B6,2C8,2C9,2C19,2D6,2E1,3A4 |
In addition it is possible to input a SMILES string from the command line.
|
1 2 |
MacPro:~ Chris$ java -jar /Users/Chris/Downloads/Leon_Ti-cypreact-cea5eab4cbd2/CypReactBundle/cypreact.jar /Users/Chris/Downloads/Leon_Ti-cypreact-cea5eab4cbd2/CypReactBundle/ SMILES='c1ccccc1NCC' /Users/Chris/Desktop/SampleFiles/emendmetab.sdf 1A2,2A6,2B6,2C8,2C19,2E1,3A4,2D6 |
If we look at Emend
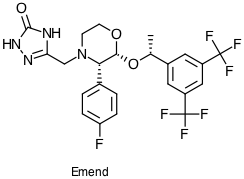
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 |
MacPro:~ Chris$ java -jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/cypreact.jar /Users/Chris/Downloads/Leon_Ti-cypreact-29a582219630/CypReactBundle/ /Users/Chris/Desktop/SampleFiles/emend.sdf /Users/Chris/Desktop/SampleFiles/emendmetab.sdf 1A2,2A6,2B6,2C8,2C9,2C19,2D6,2E1,3A4 Processing Molecue: 1 -------------------------------------- 1A2, Mole1: R -------------------------------------- 3A4, Mole1: R -------------------------------------- 2B6, Mole1: N -------------------------------------- 2E1, Mole1: N -------------------------------------- 2C9, Mole1: R -------------------------------------- 2C19, Mole1: R -------------------------------------- 2D6, Mole1: R -------------------------------------- 2C8, Mole1: N -------------------------------------- 2A6, Mole1: N |
The output file can be opened in Vortex to see the results more clearly.

One use case might be to look at series of molecules from a virtual screen (shown below), molecules that are predicted to be metabolised by multiple Cyp P450 might be less attractive. Alternatively the results might be used to suggest which enzymes might be best to prioritise in the early screening cascade.
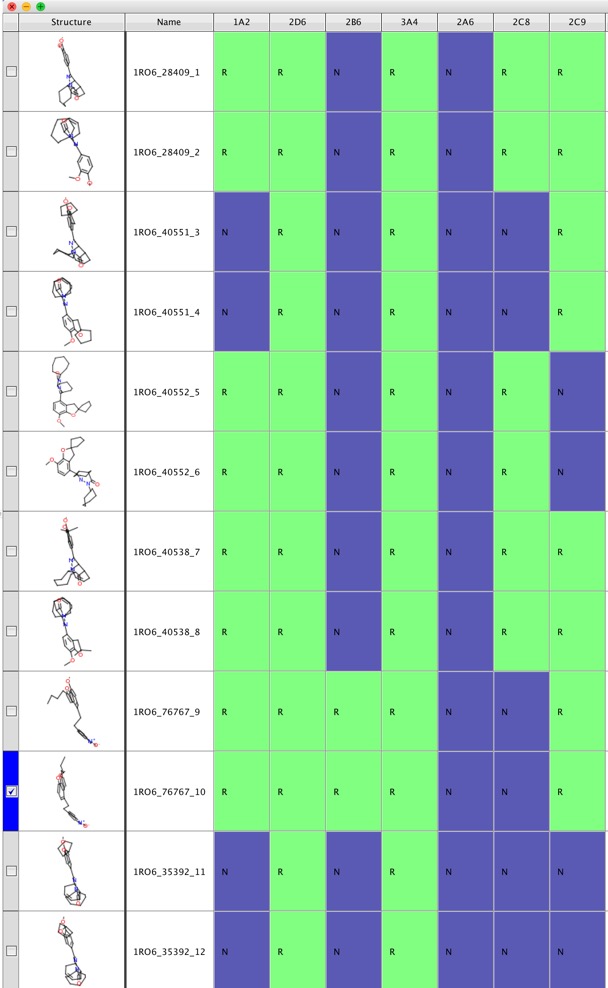
I’ve been in touch with the developers a couple of times and have been really impressed with how quickly they respond.
Last Updated 31 May 2018