ChemDraw available from Cambridgesoft has long been the perhaps the preferred package for drawing chemical structures for publication quality graphics, it also has been used as the drawing package for database queries and electronic notebooks. As the package has developed enhancements such as NMR spectra prediction, TLC plate tools, molecular and physicochemical property calculations and structure naming have been added. With the new version (Version 11) perhaps the first suprise is the name change to reflect the increased emphasis on the biological drawing features of BioDraw.

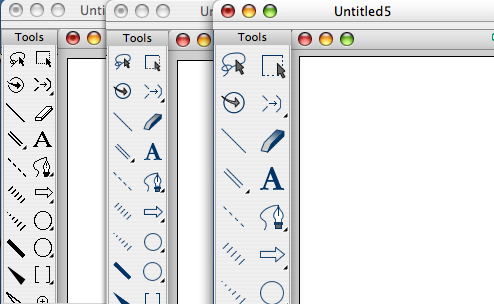
On starting up ChemBioDraw the new theme style to the tools bar is apparent (shown in the graphic below), on the left is the original tools bar, in the centre is the new default theme and on the right a new larger format. The larger format display is actually very useful for those of us who’s failing eyesight suffers when using the new large high resolution monitors. You can switch between the different themes using an option in the preferences. Using the new style highlighted tools are indicated by a darkened embossed box, multichoice tools by line square. The toolbars are actually defined by a set of XML files that are installed with ChemBioDraw. By editing the XML files, you can modify the toolbar for the way you work. You can remove, rename, and resize buttons; change button icons, or move them from one toolbar to another. You can even create or delete a toolbar altogether. The icons are stored as png files and can again be modified or replaced.
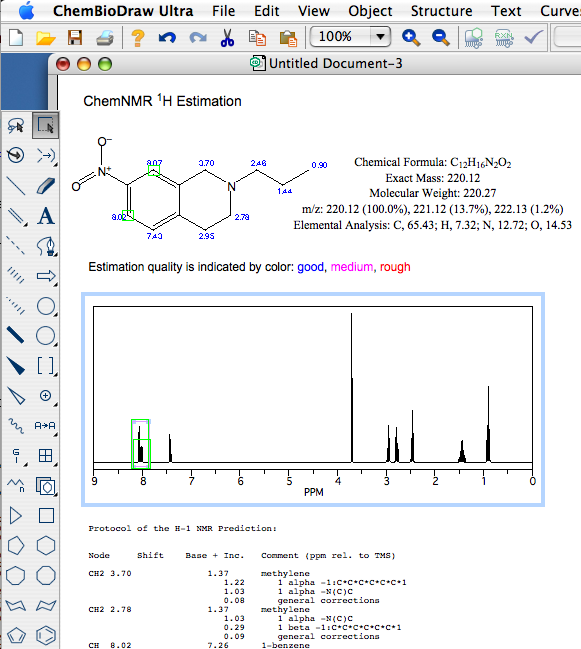
For the chemist the ChemDraw tools are very much the same, the drawing capabilities are excellent and the ability to generate and add analytical data remain unchanged. The ability to predict and proton or carbon NMR spectra are very useful with the hot linking between spectra and structure being a great way to work through peak assignments. Whilst ChemBioDraw will open JCAMP spectra files and display them it is of very limited utility. The Windows version includes a copy of the no longer updated “MestReC Std” (it is actually MestReC Lite), it is perhaps a shame that Cambridgesoft has not included a similar tool for Macintosh users perhaps something like access to iNMR Reader or similar? The main tool bar is pretty much unchanged, the only difference is moving the table and tlc plate tools to and “Advanced tools” button, this has allowed the introduction of “Sequence Tools”.
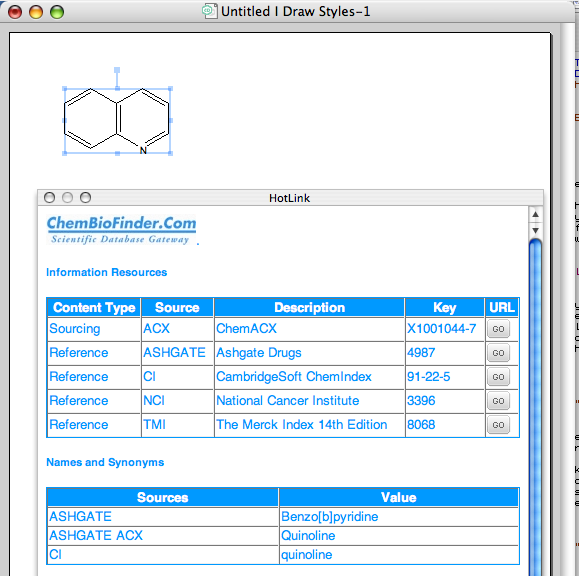
A rather nice new feature is the Database Hotlink window. To use the Show Database HotLink Window feature in ChemDraw first select from the “View” menu and choose “Show Database HotLink Window”, this opens an interactive link to online databases. Draw your structure and the results from the search on the Database Gateway web service will be displayed in the Database HotLink Window If you modify your structure the results in the Database HotLink Window will be updated to show the results of the search for the new structure If you have more than one structure shown on the page you will need to select the structure you want results for.
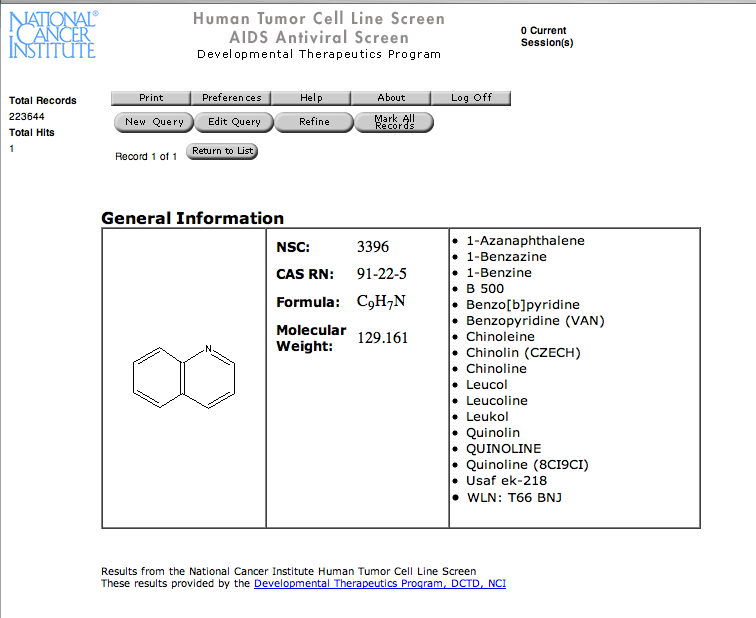
If you click on the URL button in the HotLink window your web browser will open with the relevant information, you will need to have registered for some of the databases. In this case it gives data from the National Cancer Institute screening information. Unfortuantely I don’t think it is possible to customise the hotlinking to point to your own databases.
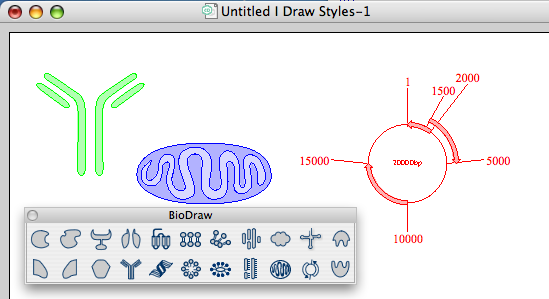
The biological drawing features have been enhanced and the BioDraw tool bar now includes several biological pathway drawing elements including membranes, DNA, enzymes, receptors, and reaction arrows. tRNA, Ribosomes, Helix Proteins, Golgi Bodies, G-Proteins, Immunoglobins, Mitochondrion, new Freehand Pen Tool, Annotation, a Plasmid mapping tool and amino acid and DNA sequence tools are also included.
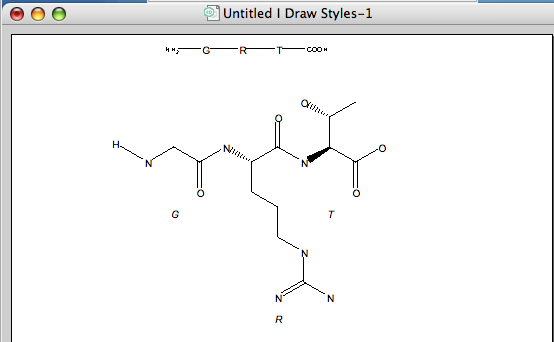
The expanding names feature is particularly useful for drawing biological molecules, you can use the sequence tool to enter a peptide using the single letter codes them use the “Expand Labels” feature from the “Structure” menu to generate the chemical structure. You can do the same thing with both DNA and RNA sequences.
If you have any Applescripts that work with ChemDraw then you can put them in the “ChemDraw Items” folder, the next time you restart ChemBioDraw a “Scripts” menu will appear in the top menu bar. It would have been nice if Cambridgesoft included a scripts folder in the ChemDraw items folder, maybe next time.